Multivariate Bijections
Background
The fundamental idea of normalizing flows also applies to multivariate random variables, and this is where its value is clearly seen - representing complex high-dimensional distributions. In this case, a simple multivariate source of noise, for example a standard i.i.d. normal distribution, , is passed through a vector-valued bijection, , to produce the more complex transformed variable .
Sampling is again trivial and involves evaluation of the forward pass of . We can score using the multivariate substitution rule of integral calculus,
where denotes the Jacobian matrix of . Equating the last two lines we get,
Inituitively, this equation says that the density of is equal to the density at the corresponding point in plus a term that corrects for the warp in volume around an infinitesimally small volume around caused by the transformation. For instance, in -dimensions, the geometric interpretation of the absolute value of the determinant of a Jacobian is that it represents the area of a parallelogram with edges defined by the columns of the Jacobian. In -dimensions, the geometric interpretation of the absolute value of the determinant Jacobian is that is represents the hyper-volume of a parallelepiped with edges defined by the columns of the Jacobian (see a calculus reference such as [7] for more details).
Similar to the univariate case, we can compose such bijective transformations to produce even more complex distributions. By an inductive argument, if we have transforms , then the log-density of the transformed variable is
where we've defined , for convenience of notation.
The main challenge is in designing parametrizable multivariate bijections that have closed form expressions for both and , a tractable Jacobian whose calculation scales with or rather than , and can express a flexible class of functions.
Multivariate Bijectors
In this section, we show how to use bij.SplineAutoregressive to learn the bivariate toy distribution from our running example. Making a simple change we can represent bivariate distributions of the form, :
dist_x = torch.distributions.Independent(
torch.distributions.Normal(torch.zeros(2), torch.ones(2)),
1
)
bijector = bij.SplineAutoregressive()
dist_y = dist.Flow(dist_x, bijector)
The bij.SplineAutoregressive bijector extends bij.Spline so that the spline parameters are the output of an autoregressive neural network. See [durkan2019neural] and [germain2015made] for more details.
Similarly to before, we train this distribution on the toy dataset and plot the results:
dataset = torch.tensor(X, dtype=torch.float)
optimizer = torch.optim.Adam(spline_transform.parameters(), lr=5e-3)
for step in range(steps):
optimizer.zero_grad()
loss = -dist_y.log_prob(dataset).mean()
loss.backward()
optimizer.step()
if step % 500 == 0:
print('step: {}, loss: {}'.format(step, loss.item()))
step: 0, loss: 8.446191787719727
step: 500, loss: 2.0197808742523193
step: 1000, loss: 1.794958472251892
step: 1500, loss: 1.73616361618042
step: 2000, loss: 1.7254879474639893
step: 2500, loss: 1.691617488861084
step: 3000, loss: 1.679549217224121
step: 3500, loss: 1.6967085599899292
step: 4000, loss: 1.6723777055740356
step: 4500, loss: 1.6505967378616333
step: 5000, loss: 1.8024061918258667
X_flow = dist_y.sample(torch.Size([1000,])).detach().numpy()
plt.title(r'Joint Distribution')
plt.xlabel(r'$x_1$')
plt.ylabel(r'$x_2$')
plt.scatter(X[:,0], X[:,1], label='data', alpha=0.5)
plt.scatter(X_flow[:,0], X_flow[:,1], color='firebrick', label='flow', alpha=0.5)
plt.legend()
plt.show()

plt.subplot(1, 2, 1)
sns.distplot(X[:,0], hist=False, kde=True,
bins=None,
hist_kws={'edgecolor':'black'},
kde_kws={'linewidth': 2},
label='data')
sns.distplot(X_flow[:,0], hist=False, kde=True,
bins=None, color='firebrick',
hist_kws={'edgecolor':'black'},
kde_kws={'linewidth': 2},
label='flow')
plt.title(r'$p(x_1)$')
plt.subplot(1, 2, 2)
sns.distplot(X[:,1], hist=False, kde=True,
bins=None,
hist_kws={'edgecolor':'black'},
kde_kws={'linewidth': 2},
label='data')
sns.distplot(X_flow[:,1], hist=False, kde=True,
bins=None, color='firebrick',
hist_kws={'edgecolor':'black'},
kde_kws={'linewidth': 2},
label='flow')
plt.title(r'$p(x_2)$')
plt.show()
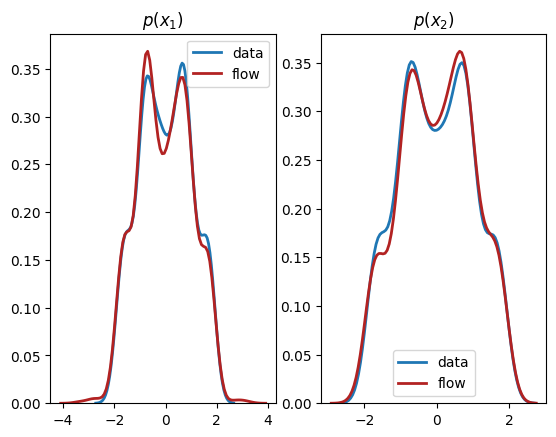
We see from the output that this normalizing flow has successfully learnt both the univariate marginals and the bivariate distribution.